Getting Started
For an outline of MLJ's goals and features, see About MLJ.
This page introduces some MLJ basics, assuming some familiarity with machine learning. For a complete list of other MLJ learning resources, see Learning MLJ.
MLJ collects together the functionality provided by mutliple packages. To learn how to install components separately, run using MLJ; @doc MLJ.
This section introduces only the most basic MLJ operations and concepts. It assumes MLJ has been successfully installed. See Installation if this is not the case.
Choosing and evaluating a model
The following code loads Fisher's famous iris data set as a named tuple of column vectors:
julia> using MLJjulia> iris = load_iris();julia> selectrows(iris, 1:3) |> pretty┌──────────────┬─────────────┬──────────────┬─────────────┬──────────────────────────────────┐ │ sepal_length │ sepal_width │ petal_length │ petal_width │ target │ │ Float64 │ Float64 │ Float64 │ Float64 │ CategoricalValue{String, UInt32} │ │ Continuous │ Continuous │ Continuous │ Continuous │ Multiclass{3} │ ├──────────────┼─────────────┼──────────────┼─────────────┼──────────────────────────────────┤ │ 5.1 │ 3.5 │ 1.4 │ 0.2 │ setosa │ │ 4.9 │ 3.0 │ 1.4 │ 0.2 │ setosa │ │ 4.7 │ 3.2 │ 1.3 │ 0.2 │ setosa │ └──────────────┴─────────────┴──────────────┴─────────────┴──────────────────────────────────┘julia> schema(iris)┌──────────────┬───────────────┬──────────────────────────────────┐ │ names │ scitypes │ types │ ├──────────────┼───────────────┼──────────────────────────────────┤ │ sepal_length │ Continuous │ Float64 │ │ sepal_width │ Continuous │ Float64 │ │ petal_length │ Continuous │ Float64 │ │ petal_width │ Continuous │ Float64 │ │ target │ Multiclass{3} │ CategoricalValue{String, UInt32} │ └──────────────┴───────────────┴──────────────────────────────────┘
Because this data format is compatible with Tables.jl (and satisfies Tables.istable(iris) == true) many MLJ methods (such as selectrows, pretty and schema used above) as well as many MLJ models can work with it. However, as most new users are already familiar with the access methods particular to DataFrames (also compatible with Tables.jl) we'll put our data into that format here:
import DataFrames
iris = DataFrames.DataFrame(iris);Next, let's split the data "horizontally" into input and target parts, and specify an RNG seed, to force observations to be shuffled:
julia> y, X = unpack(iris, ==(:target); rng=123);julia> first(X, 3) |> pretty┌──────────────┬─────────────┬──────────────┬─────────────┐ │ sepal_length │ sepal_width │ petal_length │ petal_width │ │ Float64 │ Float64 │ Float64 │ Float64 │ │ Continuous │ Continuous │ Continuous │ Continuous │ ├──────────────┼─────────────┼──────────────┼─────────────┤ │ 6.7 │ 3.3 │ 5.7 │ 2.1 │ │ 5.7 │ 2.8 │ 4.1 │ 1.3 │ │ 7.2 │ 3.0 │ 5.8 │ 1.6 │ └──────────────┴─────────────┴──────────────┴─────────────┘
This call to unpack splits off any column with name == to :target into something called y, and all the remaining columns into X.
To list all models available in MLJ's model registry do models(). Listing the models compatible with the present data:
julia> models(matching(X,y))54-element Vector{NamedTuple{(:name, :package_name, :is_supervised, :abstract_type, :deep_properties, :docstring, :fit_data_scitype, :human_name, :hyperparameter_ranges, :hyperparameter_types, :hyperparameters, :implemented_methods, :inverse_transform_scitype, :is_pure_julia, :is_wrapper, :iteration_parameter, :load_path, :package_license, :package_url, :package_uuid, :predict_scitype, :prediction_type, :reporting_operations, :reports_feature_importances, :supports_class_weights, :supports_online, :supports_training_losses, :supports_weights, :transform_scitype, :input_scitype, :target_scitype, :output_scitype)}}: (name = AdaBoostClassifier, package_name = MLJScikitLearnInterface, ... ) (name = AdaBoostStumpClassifier, package_name = DecisionTree, ... ) (name = BaggingClassifier, package_name = MLJScikitLearnInterface, ... ) (name = BayesianLDA, package_name = MLJScikitLearnInterface, ... ) (name = BayesianLDA, package_name = MultivariateStats, ... ) (name = BayesianQDA, package_name = MLJScikitLearnInterface, ... ) (name = BayesianSubspaceLDA, package_name = MultivariateStats, ... ) (name = CatBoostClassifier, package_name = CatBoost, ... ) (name = ConstantClassifier, package_name = MLJModels, ... ) (name = DecisionTreeClassifier, package_name = BetaML, ... ) ⋮ (name = SGDClassifier, package_name = MLJScikitLearnInterface, ... ) (name = SVC, package_name = LIBSVM, ... ) (name = SVMClassifier, package_name = MLJScikitLearnInterface, ... ) (name = SVMLinearClassifier, package_name = MLJScikitLearnInterface, ... ) (name = SVMNuClassifier, package_name = MLJScikitLearnInterface, ... ) (name = StableForestClassifier, package_name = SIRUS, ... ) (name = StableRulesClassifier, package_name = SIRUS, ... ) (name = SubspaceLDA, package_name = MultivariateStats, ... ) (name = XGBoostClassifier, package_name = XGBoost, ... )
In MLJ a model is a struct storing the hyperparameters of the learning algorithm indicated by the struct name (and nothing else). For common problems matching data to models, see Model Search and Preparing Data.
To see the documentation for DecisionTreeClassifier (without loading its defining code) do
doc("DecisionTreeClassifier", pkg="DecisionTree")Assuming the MLJDecisionTreeInterface.jl package is in your load path (see Installation) we can use @load to import the DecisionTreeClassifier model type, which we will bind to Tree:
julia> Tree = @load DecisionTreeClassifier pkg=DecisionTree[ Info: For silent loading, specify `verbosity=0`. import MLJDecisionTreeInterface ✔ MLJDecisionTreeInterface.DecisionTreeClassifier
(In this case, we need to specify pkg=... because multiple packages provide a model type with the name DecisionTreeClassifier.) Now we can instantiate a model with default hyperparameters:
julia> tree = Tree()DecisionTreeClassifier( max_depth = -1, min_samples_leaf = 1, min_samples_split = 2, min_purity_increase = 0.0, n_subfeatures = 0, post_prune = false, merge_purity_threshold = 1.0, display_depth = 5, feature_importance = :impurity, rng = Random._GLOBAL_RNG())
Important: DecisionTree.jl and most other packages implementing machine learning algorithms for use in MLJ are not MLJ dependencies. If such a package is not in your load path you will receive an error explaining how to add the package to your current environment. Alternatively, you can use the interactive macro @iload. For more on importing model types, see Loading Model Code.
Once instantiated, a model's performance can be evaluated with the evaluate method. Our classifier is a probabilistic predictor (check prediction_type(tree) == :probabilistic) which means we can specify a probabilistic measure (metric) like log_loss, as well deterministic measures like accuracy (which are applied after computing the mode of each prediction):
julia> evaluate(tree, X, y, resampling=CV(shuffle=true), measures=[log_loss, accuracy], verbosity=0)PerformanceEvaluation object with these fields: model, measure, operation, measurement, per_fold, per_observation, fitted_params_per_fold, report_per_fold, train_test_rows, resampling, repeats Extract: ┌──────────────────────┬──────────────┬─────────────┬─────────┬───────────────── │ measure │ operation │ measurement │ 1.96*SE │ per_fold ⋯ ├──────────────────────┼──────────────┼─────────────┼─────────┼───────────────── │ LogLoss( │ predict │ 1.92 │ 1.53 │ [1.44, 2.88, 2 ⋯ │ tol = 2.22045e-16) │ │ │ │ ⋯ │ Accuracy() │ predict_mode │ 0.947 │ 0.0425 │ [0.96, 0.92, 1 ⋯ └──────────────────────┴──────────────┴─────────────┴─────────┴───────────────── 1 column omitted
Under the hood, evaluate calls lower level functions predict or predict_mode according to the type of measure, as shown in the output. We shall call these operations directly below.
For more on performance evaluation, see Evaluating Model Performance for details.
A preview of data type specification in MLJ
The target y above is a categorical vector, which is appropriate because our model is a decision tree classifier:
julia> typeof(y)CategoricalVector{String, UInt32, String, CategoricalValue{String, UInt32}, Union{}} (alias for CategoricalArray{String, 1, UInt32, String, CategoricalValue{String, UInt32}, Union{}})
However, MLJ models do not prescribe the machine types for the data they operate on. Rather, they specify a scientific type, which refers to the way data is to be interpreted, as opposed to how it is encoded:
julia> target_scitype(tree)AbstractVector{<:Finite} (alias for AbstractArray{<:Finite, 1})
Here Finite is an example of a "scalar" scientific type with two subtypes:
julia> subtypes(Finite)2-element Vector{Any}: Multiclass OrderedFactor
We use the scitype function to check how MLJ is going to interpret given data. Our choice of encoding for y works for DecisionTreeClassifier, because we have:
julia> scitype(y)AbstractVector{Multiclass{3}} (alias for AbstractArray{Multiclass{3}, 1})
and Multiclass{3} <: Finite. If we would encode with integers instead, we obtain:
julia> yint = int.(y);julia> scitype(yint)AbstractVector{Count} (alias for AbstractArray{Count, 1})
and using yint in place of y in classification problems will fail. See also Working with Categorical Data.
For more on scientific types, see Data containers and scientific types below.
Fit and predict
To illustrate MLJ's fit and predict interface, let's perform our performance evaluations by hand, but using a simple holdout set, instead of cross-validation.
Wrapping the model in data creates a machine which will store training outcomes:
julia> mach = machine(tree, X, y)untrained Machine; caches model-specific representations of data model: DecisionTreeClassifier(max_depth = -1, …) args: 1: Source @967 ⏎ Table{AbstractVector{Continuous}} 2: Source @156 ⏎ AbstractVector{Multiclass{3}}
Training and testing on a hold-out set:
julia> train, test = partition(eachindex(y), 0.7); # 70:30 splitjulia> fit!(mach, rows=train);[ Info: Training machine(DecisionTreeClassifier(max_depth = -1, …), …).julia> yhat = predict(mach, X[test,:]);julia> yhat[3:5]3-element UnivariateFiniteVector{Multiclass{3}, String, UInt32, Float64}: UnivariateFinite{Multiclass{3}}(setosa=>1.0, versicolor=>0.0, virginica=>0.0) UnivariateFinite{Multiclass{3}}(setosa=>0.0, versicolor=>0.0, virginica=>1.0) UnivariateFinite{Multiclass{3}}(setosa=>0.0, versicolor=>0.0, virginica=>1.0)julia> log_loss(yhat, y[test])2.4029102259411435
Note that log_loss and cross_entropy are aliases for LogLoss() (which can be passed an optional keyword parameter, as in LogLoss(tol=0.001)). For a list of all losses and scores, and their aliases, run measures().
Notice that yhat is a vector of Distribution objects, because DecisionTreeClassifier makes probabilistic predictions. The methods of the Distributions.jl package can be applied to such distributions:
julia> broadcast(pdf, yhat[3:5], "virginica") # predicted probabilities of virginica3-element Vector{Float64}: 0.0 1.0 1.0julia> broadcast(pdf, yhat, y[test])[3:5] # predicted probability of observed class3-element Vector{Float64}: 1.0 1.0 1.0julia> mode.(yhat[3:5])3-element CategoricalArray{String,1,UInt32}: "setosa" "virginica" "virginica"
Or, one can explicitly get modes by using predict_mode instead of predict:
julia> predict_mode(mach, X[test[3:5],:])3-element CategoricalArray{String,1,UInt32}: "setosa" "virginica" "virginica"
Finally, we note that pdf() is overloaded to allow the retrieval of probabilities for all levels at once:
julia> L = levels(y)3-element Vector{String}: "setosa" "versicolor" "virginica"julia> pdf(yhat[3:5], L)3×3 Matrix{Float64}: 1.0 0.0 0.0 0.0 0.0 1.0 0.0 0.0 1.0
Unsupervised models have a transform method instead of predict, and may optionally implement an inverse_transform method:
julia> v = Float64[1, 2, 3, 4]4-element Vector{Float64}: 1.0 2.0 3.0 4.0julia> stand = Standardizer() # this type is built-inStandardizer( features = Symbol[], ignore = false, ordered_factor = false, count = false)julia> mach2 = machine(stand, v)untrained Machine; caches model-specific representations of data model: Standardizer(features = Symbol[], …) args: 1: Source @585 ⏎ AbstractVector{Continuous}julia> fit!(mach2)[ Info: Training machine(Standardizer(features = Symbol[], …), …). trained Machine; caches model-specific representations of data model: Standardizer(features = Symbol[], …) args: 1: Source @585 ⏎ AbstractVector{Continuous}julia> w = transform(mach2, v)4-element Vector{Float64}: -1.161895003862225 -0.3872983346207417 0.3872983346207417 1.161895003862225julia> inverse_transform(mach2, w)4-element Vector{Float64}: 1.0 2.0 3.0 4.0
Machines have an internal state which allows them to avoid redundant calculations when retrained, in certain conditions - for example when increasing the number of trees in a random forest, or the number of epochs in a neural network. The machine-building syntax also anticipates a more general syntax for composing multiple models, an advanced feature explained in Learning Networks.
There is a version of evaluate for machines as well as models. This time we'll use a simple holdout strategy as above. (An exclamation point is added to the method name because machines are generally mutated when trained.)
julia> evaluate!(mach, resampling=Holdout(fraction_train=0.7), measures=[log_loss, accuracy], verbosity=0)PerformanceEvaluation object with these fields: model, measure, operation, measurement, per_fold, per_observation, fitted_params_per_fold, report_per_fold, train_test_rows, resampling, repeats Extract: ┌──────────────────────┬──────────────┬─────────────┬──────────┐ │ measure │ operation │ measurement │ per_fold │ ├──────────────────────┼──────────────┼─────────────┼──────────┤ │ LogLoss( │ predict │ 2.4 │ [2.4] │ │ tol = 2.22045e-16) │ │ │ │ │ Accuracy() │ predict_mode │ 0.933 │ [0.933] │ └──────────────────────┴──────────────┴─────────────┴──────────┘
Changing a hyperparameter and re-evaluating:
julia> tree.max_depth = 33julia> evaluate!(mach, resampling=Holdout(fraction_train=0.7), measures=[log_loss, accuracy], verbosity=0)PerformanceEvaluation object with these fields: model, measure, operation, measurement, per_fold, per_observation, fitted_params_per_fold, report_per_fold, train_test_rows, resampling, repeats Extract: ┌──────────────────────┬──────────────┬─────────────┬──────────┐ │ measure │ operation │ measurement │ per_fold │ ├──────────────────────┼──────────────┼─────────────┼──────────┤ │ LogLoss( │ predict │ 1.61 │ [1.61] │ │ tol = 2.22045e-16) │ │ │ │ │ Accuracy() │ predict_mode │ 0.956 │ [0.956] │ └──────────────────────┴──────────────┴─────────────┴──────────┘
Next steps
For next steps, consult the Learning MLJ section. At the least, we recommned you read the remainder of this page before considering serious use of MLJ.
Data containers and scientific types
The MLJ user should acquaint themselves with some basic assumptions about the form of data expected by MLJ, as outlined below. The basic machine constructors look like this (see also Constructing machines):
machine(model::Unsupervised, X)
machine(model::Supervised, X, y)Each supervised model in MLJ declares the permitted scientific type of the inputs X and targets y that can be bound to it in the first constructor above, rather than specifying specific machine types (such as Array{Float32, 2}). Similar remarks apply to the input X of an unsupervised model.
Scientific types are julia types defined in the package ScientificTypesBase.jl; the package ScientificTypes.jl implements the particular convention used in the MLJ universe for assigning a specific scientific type (interpretation) to each julia object (see the scitype examples below).
The basic "scalar" scientific types are Continuous, Multiclass{N}, OrderedFactor{N}, Count and Textual. Missing and Nothing are also considered scientific types. Be sure you read Scalar scientific types below to guarantee your scalar data is interpreted correctly. Tools exist to coerce the data to have the appropriate scientific type; see ScientificTypes.jl or run ?coerce for details.
Additionally, most data containers - such as tuples, vectors, matrices and tables - have a scientific type parameterized by scitype of the elements they contain.
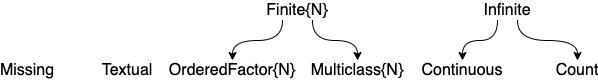
Figure 1. Part of the scientific type hierarchy in ScientificTypesBase.jl.
julia> scitype(4.6)Continuousjulia> scitype(42)Countjulia> x1 = coerce(["yes", "no", "yes", "maybe"], Multiclass);julia> scitype(x1)AbstractVector{Multiclass{3}} (alias for AbstractArray{Multiclass{3}, 1})julia> X = (x1=x1, x2=rand(4), x3=rand(4)) # a "column table"(x1 = CategoricalValue{String, UInt32}["yes", "no", "yes", "maybe"], x2 = [0.17296982732153476, 0.4311265688549152, 0.9164218371622808, 0.0029817110637152533], x3 = [0.7422777179776697, 0.5491931062469285, 0.05065936857102282, 0.14872233376483412],)julia> scitype(X)Table{Union{AbstractVector{Continuous}, AbstractVector{Multiclass{3}}}}
Two-dimensional data
Generally, two-dimensional data in MLJ is expected to be tabular. All data containers X compatible with the Tables.jl interface and sastisfying Tables.istable(X) == true (most of the formats in this list) have the scientific type Table{K}, where K depends on the scientific types of the columns, which can be individually inspected using schema:
julia> schema(X)┌───────┬───────────────┬──────────────────────────────────┐ │ names │ scitypes │ types │ ├───────┼───────────────┼──────────────────────────────────┤ │ x1 │ Multiclass{3} │ CategoricalValue{String, UInt32} │ │ x2 │ Continuous │ Float64 │ │ x3 │ Continuous │ Float64 │ └───────┴───────────────┴──────────────────────────────────┘
Matrix data
MLJ models expecting a table do not generally accept a matrix instead. However, a matrix can be wrapped as a table, using MLJ.table:
julia> matrix_table = MLJ.table(rand(2,3));julia> schema(matrix_table)┌───────┬────────────┬─────────┐ │ names │ scitypes │ types │ ├───────┼────────────┼─────────┤ │ x1 │ Continuous │ Float64 │ │ x2 │ Continuous │ Float64 │ │ x3 │ Continuous │ Float64 │ └───────┴────────────┴─────────┘
The matrix is not copied, only wrapped. To manifest a table as a matrix, use MLJ.matrix.
Observations correspond to rows, not columns
When supplying models with matrices, or wrapping them in tables, each row should correspond to a different observation. That is, the matrix should be n x p, where n is the number of observations and p the number of features. However, some models may perform better if supplied the adjoint of a p x n matrix instead, and observation resampling is always more efficient in this case.
Inputs
Since an MLJ model only specifies the scientific type of data, if that type is Table - which is the case for the majority of MLJ models - then any Tables.jl container X is permitted, so long as Tables.istable(X) == true.
Specifically, the requirement for an arbitrary model's input is scitype(X) <: input_scitype(model).
Targets
The target y expected by MLJ models is generally an AbstractVector. A multivariate target y will generally be a table.
Specifically, the type requirement for a model target is scitype(y) <: target_scitype(model).
Querying a model for acceptable data types
Given a model instance, one can inspect the admissible scientific types of its input and target, and without loading the code defining the model;
julia> i = info("DecisionTreeClassifier", pkg="DecisionTree")(name = "DecisionTreeClassifier", package_name = "DecisionTree", is_supervised = true, abstract_type = Probabilistic, deep_properties = (), docstring = "```\nDecisionTreeClassifier\n```\n\nA model type for c...", fit_data_scitype = Tuple{Table{<:Union{AbstractVector{<:Continuous}, AbstractVector{<:Count}, AbstractVector{<:OrderedFactor}}}, AbstractVector{<:Finite}}, human_name = "CART decision tree classifier", hyperparameter_ranges = (nothing, nothing, nothing, nothing, nothing, nothing, nothing, nothing, nothing, nothing), hyperparameter_types = ("Int64", "Int64", "Int64", "Float64", "Int64", "Bool", "Float64", "Int64", "Symbol", "Union{Integer, Random.AbstractRNG}"), hyperparameters = (:max_depth, :min_samples_leaf, :min_samples_split, :min_purity_increase, :n_subfeatures, :post_prune, :merge_purity_threshold, :display_depth, :feature_importance, :rng), implemented_methods = [:clean!, :fit, :fitted_params, :predict, :reformat, :selectrows, :feature_importances], inverse_transform_scitype = Unknown, is_pure_julia = true, is_wrapper = false, iteration_parameter = nothing, load_path = "MLJDecisionTreeInterface.DecisionTreeClassifier", package_license = "MIT", package_url = "https://github.com/bensadeghi/DecisionTree.jl", package_uuid = "7806a523-6efd-50cb-b5f6-3fa6f1930dbb", predict_scitype = AbstractVector{ScientificTypesBase.Density{_s25} where _s25<:Finite}, prediction_type = :probabilistic, reporting_operations = (), reports_feature_importances = true, supports_class_weights = false, supports_online = false, supports_training_losses = false, supports_weights = false, transform_scitype = Unknown, input_scitype = Table{<:Union{AbstractVector{<:Continuous}, AbstractVector{<:Count}, AbstractVector{<:OrderedFactor}}}, target_scitype = AbstractVector{<:Finite}, output_scitype = Unknown)julia> i.input_scitypeTable{<:Union{AbstractVector{<:Continuous}, AbstractVector{<:Count}, AbstractVector{<:OrderedFactor}}}julia> i.target_scitypeAbstractVector{<:Finite} (alias for AbstractArray{<:Finite, 1})
This output indicates that any table with Continuous, Count or OrderedFactor columns is acceptable as the input X, and that any vector with element scitype <: Finite is acceptable as the target y.
For more on matching models to data, see Model Search.
Scalar scientific types
Models in MLJ will always apply the MLJ convention described in ScientificTypes.jl to decide how to interpret the elements of your container types. Here are the key features of that convention:
Any
AbstractFloatis interpreted asContinuous.Any
Integeris interpreted asCount.Any
CategoricalValuex, is interpreted asMulticlassorOrderedFactor, depending on the value ofisordered(x).Strings andChars are not interpreted asMulticlassorOrderedFactor(they have scitypesTextualandUnknownrespectively).In particular, integers (including
Bools) cannot be used to represent categorical data. Use the precedingcoerceoperations to coerce to aFinitescitype.The scientific types of
nothingandmissingareNothingandMissing, native types we also regard as scientific.
Use coerce(v, OrderedFactor) or coerce(v, Multiclass) to coerce a vector v of integers, strings or characters to a vector with an appropriate Finite (categorical) scitype. See also Working with Categorical Data, and the ScientificTypes.jl documentation.