Data Requirements#
Before getting started with DeepSensor, it’s important to understand the package’s data requirements. Further details on what DeepSensor supports and does not support are provided below.
DeepSensor does support:
Arbitrary numbers of context sets (or data streams), each with arbitrary numbers of variables (i.e. data channels)
Spatiotemporal data and static auxiliary data
Gridded and off-the-grid data
Moving sensors
Missing data
DeepSensor does not support:
Data with irregular temporal sampling (i.e. continuous time)
DeepSensor has the following data requirements:
Data formats:
Pandas Series/DataFrame (i.e. tabular CSV data)
Xarray DataArray/Dataset (i.e. gridded NetCDF data)
Regular temporal sampling (i.e. hourly, daily, monthly, etc.), which is the same for all variables
Consistent spatiotemporal dimension names across variables (but arbitrary names are permitted, e.g.
time,lat,lon, ordate,y,x)Each variable has a unique ID, which is used to identify the variable in the model
Missing data represented by NaNs (i.e., not arbitrary values such as -9999)
Data Sources#
DeepSensor provides several functions in deepsensor.data.sources for fetching environmental data in the format expected by the package.
These are not intended as definitive data sources for DeepSensor applications, but rather as a means of demoing and getting started with the package.
The following data sources are currently supported:
GHCND: Global Historical Climatology Network Daily (GHCND) station data
ERA5: ERA5 reanalysis data
EarthEnv: EarthEnv elevation and Topographic Position Index (TPI) data at various resolutions (1 km, 5 km, 10 km, 50 km, 100 km)
GLDAS: Global Land Data Assimilation System (GLDAS) 0.25 degree resolution binary land mask
For more details on the data sources, see the API reference for the data.sources module.
Note
Some of the data downloader functions used here require additional dependencies. To run this yourself you will need to run:
pip install rioxarray
to install the rioxarray package and
pip install git+https://github.com/scott-hosking/get-station-data.git
to install the get_station_data package.
import logging
logging.captureWarnings(True)
import xarray as xr
import cartopy.crs as ccrs
import cartopy.feature as cfeature
import matplotlib.pyplot as plt
# Cache the data in root of docs/ folder, so that other notebook don't need to download it again
cache_dir = "../../.datacache"
from deepsensor.data.sources import get_ghcnd_station_data, get_era5_reanalysis_data, \
get_earthenv_auxiliary_data, get_gldas_land_mask
data_range = ("2015-06-25", "2015-06-30")
extent = "europe"
Off-the-grid station data#
Off-the-grid station data is data that is not on a regular grid, such as in-situ observations from weather stations. In DeepSensor, Pandas DataFrames are used to represent off-the-grid station data.
The variable ID is the column name
If the DataFrame has multiple columns, each column is treated as a separate variable
The indexes must be (time, x1, x2), where time is a datetime, and x1 and x2 are the spatial dimensions
There may be an arbitrary number of additional indexes after these first three indexes. This can be useful for tracking station IDs, for example.
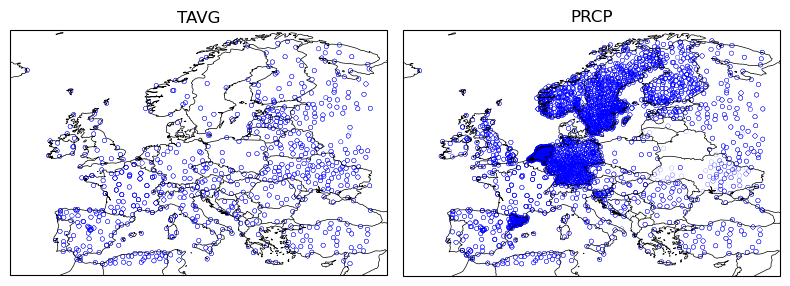
station_var_IDs = ["TAVG", "PRCP"]
station_raw_df = get_ghcnd_station_data(station_var_IDs, extent, date_range=data_range, cache=True, cache_dir=cache_dir)
station_raw_df
| PRCP | TAVG | ||||
|---|---|---|---|---|---|
| time | lat | lon | station | ||
| 2015-06-25 | 35.017 | -1.450 | AGM00060531 | 0.0 | 23.0 |
| 35.100 | -1.850 | AGE00147716 | 0.0 | 23.4 | |
| 35.117 | 36.750 | SYM00040030 | NaN | 25.4 | |
| 35.167 | 2.317 | AGM00060514 | 0.0 | 25.9 | |
| 35.200 | -0.617 | AGM00060520 | 0.0 | 24.9 | |
| ... | ... | ... | ... | ... | ... |
| 2015-06-30 | 45.933 | 7.700 | ITM00016052 | NaN | 5.7 |
| 38.367 | -0.500 | SPM00008359 | 0.0 | 27.6 | |
| 55.383 | 36.700 | RSM00027611 | 0.0 | 17.2 | |
| 59.080 | 17.860 | SWE00138750 | 0.0 | NaN | |
| 63.760 | 12.430 | SWE00140158 | 0.6 | NaN |
16922 rows × 2 columns
Spatiotemporal gridded data#
Spatiotemporal gridded data is data that is on a regular spatiotemporal grid, such as reanalysis data or satellite data. In DeepSensor, Xarray DataArrays/Datasets are used to represent gridded data.
The variable ID is the variable name in the DataArray/Dataset
The dimensions must be (time, y, x), where time is a datetime, and y and x are the spatial dimensions
era5_var_IDs = ["2m_temperature", "10m_u_component_of_wind", "10m_v_component_of_wind"]
era5_raw_ds = get_era5_reanalysis_data(era5_var_IDs, extent, date_range=data_range, cache=True, cache_dir=cache_dir)
era5_raw_ds
<xarray.Dataset>
Dimensions: (time: 6, lat: 141, lon: 221)
Coordinates:
* lat (lat) float32 70.0 69.75 69.5 ... 35.5 35.25 35.0
* lon (lon) float32 -15.0 -14.75 -14.5 ... 39.75 40.0
* time (time) datetime64[ns] 2015-06-25 ... 2015-06-30
Data variables:
2m_temperature (time, lat, lon) float32 274.7 274.8 ... 300.5
10m_u_component_of_wind (time, lat, lon) float32 5.999 5.951 ... 4.999
10m_v_component_of_wind (time, lat, lon) float32 2.87 2.746 ... -3.017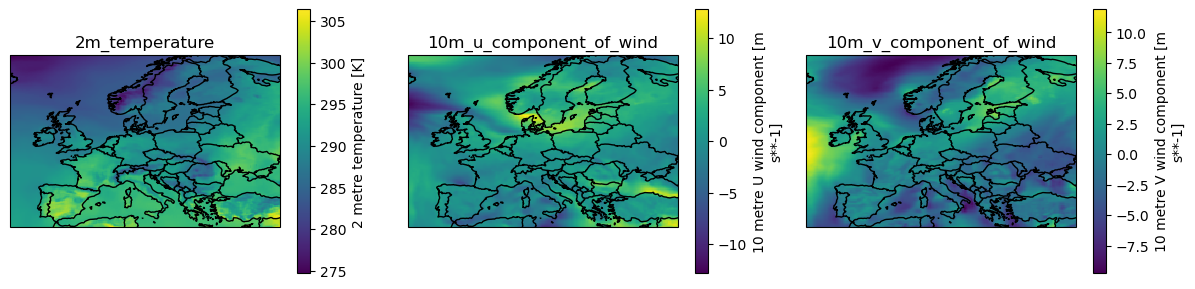
Static gridded data#
We will often want to leverage static variables in our modelling pipelines. These static variables may provide information that aid predicting our target variables. For example, topography will affect surface temperature and soil type will affect soil moisture. DeepSensor classes support these ‘auxiliary’ variables with no time dimension. Here we will download 1 km resolution elevation and Topographic Position Index (TPI) data from EarthEnv, as well as a land mask from GLDAS.
auxiliary_var_IDs = ["elevation", "tpi"]
da = get_earthenv_auxiliary_data(auxiliary_var_IDs, extent, "1KM", cache=True, cache_dir=cache_dir)
da
<xarray.Dataset>
Dimensions: (lon: 6600, lat: 4200)
Coordinates:
* lon (lon) float64 -15.0 -14.99 -14.98 -14.97 ... 39.98 39.99 40.0
* lat (lat) float64 70.0 69.99 69.98 69.97 ... 35.03 35.02 35.01 35.0
Data variables:
elevation (lat, lon) float32 0.0 0.0 0.0 0.0 ... 260.8 261.6 261.5 260.6
tpi (lat, lon) float32 0.0 0.0 0.0 0.0 ... 0.03906 0.07812 0.1641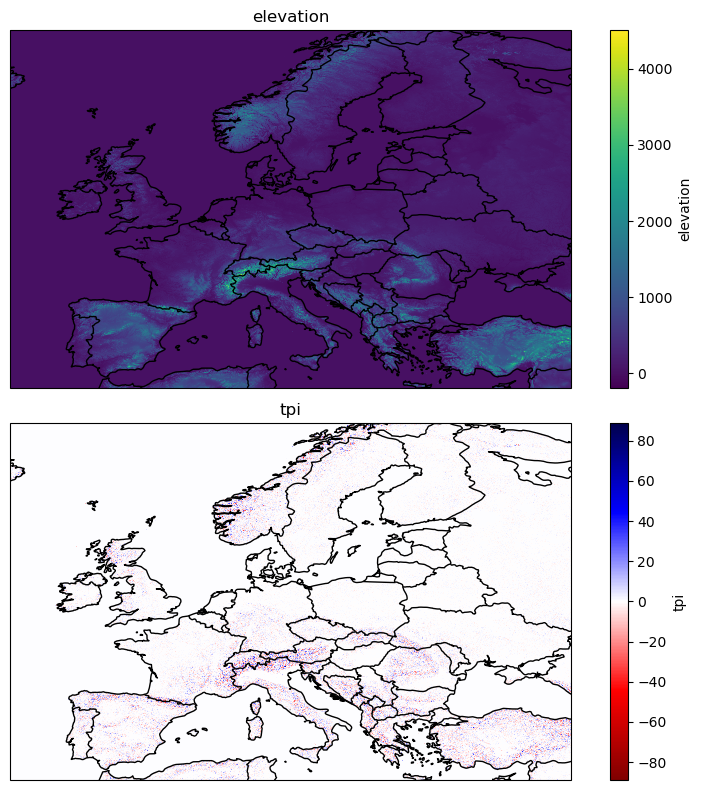
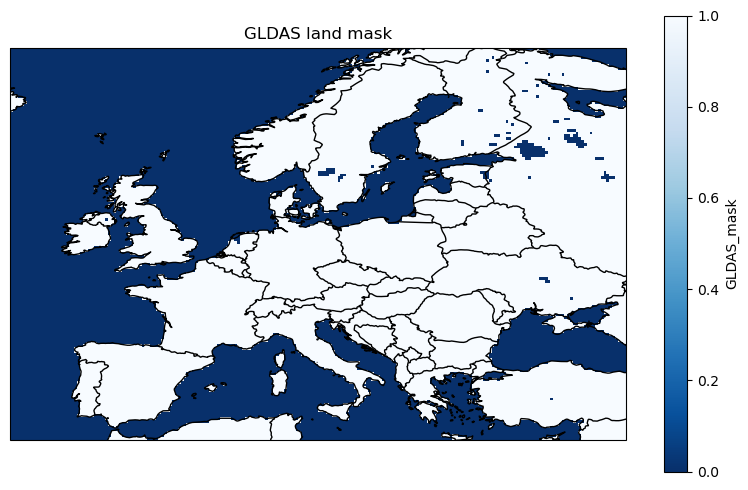
land_da = get_gldas_land_mask(extent, cache=True, cache_dir=cache_dir)
land_da
<xarray.DataArray 'GLDAS_mask' (lat: 140, lon: 220)>
array([[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
[0., 0., 0., ..., 0., 0., 0.],
...,
[0., 0., 0., ..., 1., 1., 1.],
[0., 0., 0., ..., 1., 1., 1.],
[0., 0., 0., ..., 1., 1., 1.]], dtype=float32)
Coordinates:
* lon (lon) float32 -14.88 -14.62 -14.38 -14.12 ... 39.38 39.62 39.88
* lat (lat) float32 69.88 69.62 69.38 69.12 ... 35.88 35.62 35.38 35.12