Dimensionality reduction#
Overview#
Sometimes an emulator performs better when the input and/or the output dimensionality is reduced. In this example, we show how one can specify dimensionality reduction methods to be used in combination with the emulators tested by AutoEmulate.
Reaction-Diffusion example#
We train an emulator to generate solutions to a 2D parameterized reaction-diffusion problem governed by the following partial differential equations:
where:
\( u \) and \( v \) are the concentrations of two species,
\( \beta \) and \( d \) control the reaction and diffusion terms.
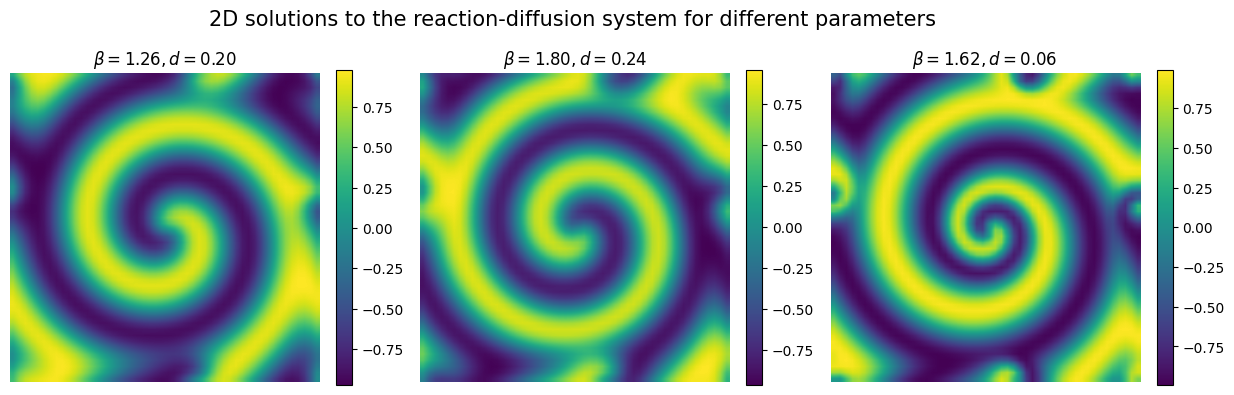
This system exhibits complex spatio-temporal dynamics such as spiral waves.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import os
import warnings
warnings.filterwarnings("ignore")
from autoemulate.simulations.reaction_diffusion import ReactionDiffusion
from autoemulate.datasets import fetch_data
save = False
train = False
1: Data generation#
Data are computed using a numerical simulator using Fourier spectral method. The simulator takes two inputs: the reaction parameter \(\beta\) and the diffusion parameter \(d\).
We sample 50 sets of inputs X using Latin Hypercube sampling and run the simulator for those inputs to get the solutions Y. Below you can either run the simulator or load data that has already been simulated.
n_samples = 50
n = 50
sim = ReactionDiffusion(n=n, T=10)
X = sim.sample_inputs(n_samples)
if train:
data_folder = "../data/reactiondiffusion1/processed"
if not os.path.exists(data_folder):
os.makedirs(data_folder)
X_file = os.path.join(data_folder, "parameters.csv")
Y_file = os.path.join(data_folder, "outputs.csv")
# Run the simulator to generate data
# Simulator returns flattened species U and V
UV, _ = sim.forward_batch(X)
# Let's consider as output the concentration of species U
m = int(UV.shape[1] / 2)
Y = UV[:, :m]
if save:
# Save the data
pd.DataFrame(X).to_csv(X_file, index=False)
pd.DataFrame(Y).to_csv(Y_file, index=False)
else:
# Load the data
X, Y = fetch_data('reactiondiffusion1')
print(f"shapes: input X: {X.shape}, output Y: {Y.shape}\n")
shapes: input X: torch.Size([50, 2]), output Y: torch.Size([50, 2500])
X and Y are matrices where each row represents one run of the simulation. In the input matrix X the two columns indicate the input parameters (reaction \(\beta\) and diffusion \(d\) parameters, respetively).
In the output matrix Y each column indicates a spatial location where the solution (i.e. the concentration of \(u\) at final time \(T=10\)) is computed.
We consider a 2D spatial grid of \(50\times 50\) points, therefore each row of Y corresponds to a 2500-dimensional vector!
Let’s now plot the simulated data to see what the reaction-diffusion pattern looks like.
plt.figure(figsize=(15,4.5))
for param in range(3):
plt.subplot(1,3,1+param)
plt.imshow(Y[param].reshape(n,n), interpolation='bilinear')
plt.axis('off')
plt.xlabel('x', fontsize=12)
plt.ylabel('y')
plt.title(r'$\beta = {:.2f}, d = {:.2f}$'.format(X[param][0], X[param][1]), fontsize=12)
plt.colorbar(fraction=0.046)
plt.suptitle('2D solutions to the reaction-diffusion system for different parameters', fontsize=15)
plt.show()
2: Fit an emulator on a subset of the data#
Below we pass a sequence of x_transforms and y_transforms to AutoEmulate. In this example we apply PCA to the simulation output and train a Gaussian Process emulator on this reduced problem space. This is often more efficient for high dimensional problems.
Note that the transforms are specified as a list of lists. This enables users to specify different transform combinations (each a list) to test with all the emulators. The transforms can be any subclass of torch.distributions.Transform - here we demonstrate how to flatten the data with ReshapeTransform. We also make use of AutoEmulate provided transforms to standardize the data before dimensionality reduction with PCA. In addition to dimensionality reduction with PCA, AutoEmulate also has an option to use Variational Autoencoder.
from autoemulate.core.compare import AutoEmulate
from autoemulate.transforms import *
from torch.distributions.transforms import ReshapeTransform
import torch
# Convert to tensors
x, y = torch.Tensor(X).float(), torch.Tensor(Y).float().reshape(-1, n, n)
# Split into train/test data
test_idx = [np.array([13, 39, 30, 45, 17, 48, 26, 25, 32, 19])]
train_idx = np.setdiff1d(np.arange(len(x)), test_idx)
# Run AutoEmulate
ae = AutoEmulate(
x[train_idx],
y[train_idx],
models=["GaussianProcessRBF"],
x_transforms_list=[[StandardizeTransform()]],
y_transforms_list=[[
ReshapeTransform((n, n), (n * n,)), # include torch transform to flatten first
StandardizeTransform(),
PCATransform(n_components=16),
StandardizeTransform()
]],
# fewer bootstraps to reduce computation time, though more uncertainty on test score
n_bootstraps=None,
log_level="error",
model_params={"posterior_predictive": True},
transformed_emulator_params={
"full_covariance": False,
"output_from_samples": False
}
)
Summarise the results across the models considered.
ae.summarise().T
| 0 | |
|---|---|
| model_name | GaussianProcessRBF |
| x_transforms | [StandardizeTransform()] |
| y_transforms | [ReshapeTransform(), StandardizeTransform(), P... |
| params | {'likelihood_cls': <class 'gpytorch.likelihood... |
| r2_test | 0.95215 |
| r2_test_std | NaN |
| rmse_test | 0.144496 |
| rmse_test_std | NaN |
| r2_train | 0.998253 |
| r2_train_std | NaN |
| rmse_train | 0.025612 |
| rmse_train_std | NaN |
Get best performing emulator and fit on all training data with best model class reinitialized and refitted on all the training data.
em = ae.fit_from_reinitialized(x[train_idx], y[train_idx])
3: Test predictions#
# Get predictions for the full dataset
y_true = y[test_idx].numpy()
y_pred_dis = em.predict(x[test_idx])
y_pred, y_var_pred = em.predict_mean_and_variance(x[test_idx])
y_pred, y_std_pred = y_pred.numpy(), y_var_pred.sqrt().numpy()
print("Prediction distribution shape:", y_pred.shape, y_std_pred.shape)
print("Mean shape:", y_pred.shape)
print("Std shape:", y_std_pred.shape)
Prediction distribution shape: (10, 50, 50) (10, 50, 50)
Mean shape: (10, 50, 50)
Std shape: (10, 50, 50)
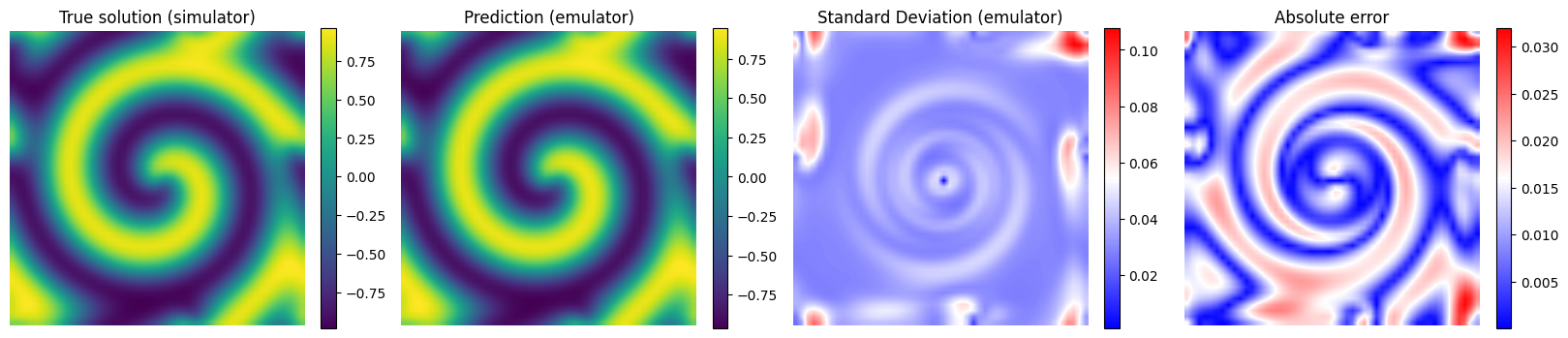
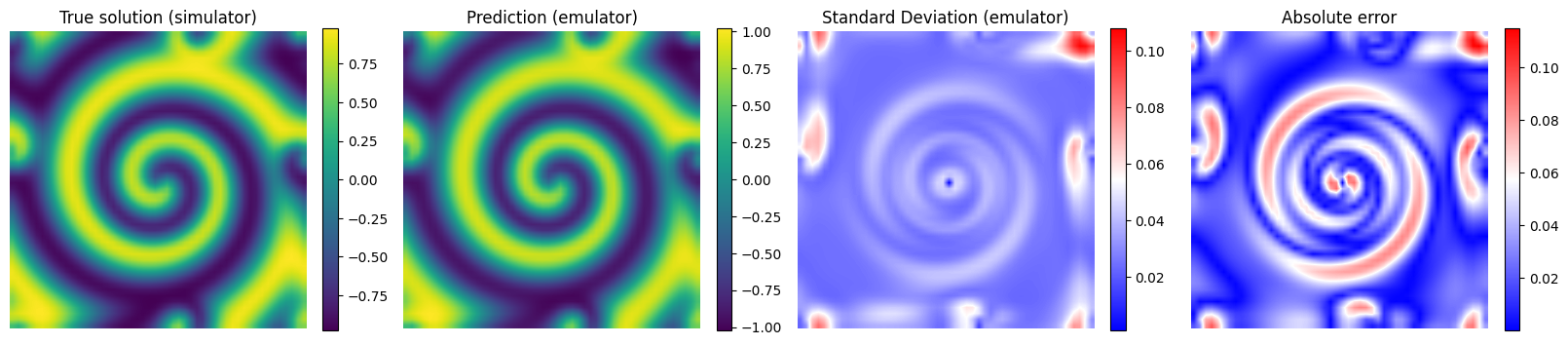
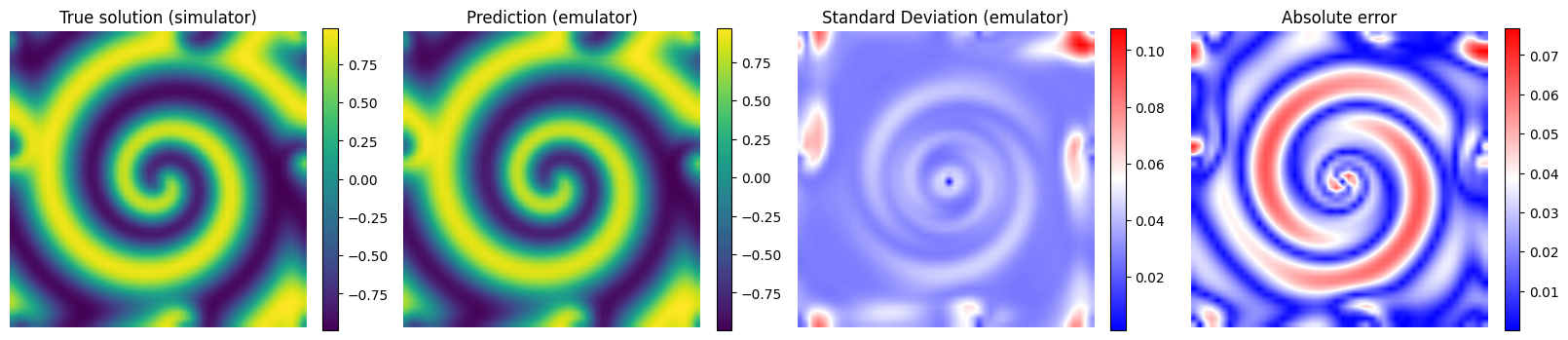
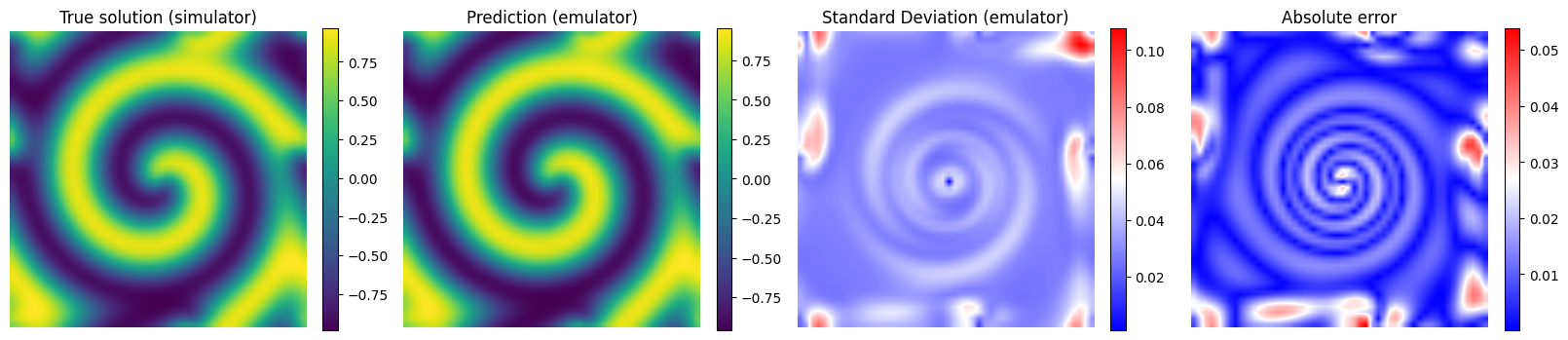
# Plot the results for some unseen (test) parameter instances
params_test = [0,1,2,3]
for param_test in params_test:
plt.figure(figsize=(20,4.5))
plt.subplot(1,4,1)
plt.imshow(y_true[param_test], interpolation='bilinear')
plt.axis('off')
plt.xlabel('x', fontsize=12)
plt.ylabel('y')
plt.title('True solution (simulator)', fontsize=12)
plt.colorbar(fraction=0.046)
plt.subplot(1,4,2)
plt.imshow(y_pred[param_test], interpolation='bilinear')
plt.axis('off')
plt.xlabel('x', fontsize=12)
plt.ylabel('y')
plt.title('Prediction (emulator)', fontsize=12)
plt.colorbar(fraction=0.046)
plt.subplot(1,4,3)
plt.imshow(y_std_pred[param_test], cmap = 'bwr', interpolation='bilinear', vmax = np.max(y_std_pred[params_test]))
plt.axis('off')
plt.xlabel('x', fontsize=12)
plt.ylabel('y')
plt.title('Standard Deviation (emulator)', fontsize=12)
plt.colorbar(fraction=0.046)
plt.subplot(1,4,4)
plt.imshow(np.abs(y_pred[param_test] - y_true[param_test]), cmap = 'bwr', interpolation='bilinear')
plt.axis('off')
plt.xlabel('x', fontsize=12)
plt.ylabel('y')
plt.title('Absolute error', fontsize=12)
plt.colorbar(fraction=0.046)
# plt.suptitle(r'Results for test parameters: $\beta = {:.2f}, d = {:.2f}$'.format(X[em.test_idxs][param_test][0], X[em.test_idxs][param_test][1]), fontsize=12)
plt.show()